A large-scale analysis of codon usage bias in 4868 bacterial genomes shows association of codon adaptation index with GC content, protein functional domains and bacterial phenotypes - ScienceDirect

SciELO - Brasil - NS1 codon usage adaptation to humans in pandemic Zika virus NS1 codon usage adaptation to humans in pandemic Zika virus

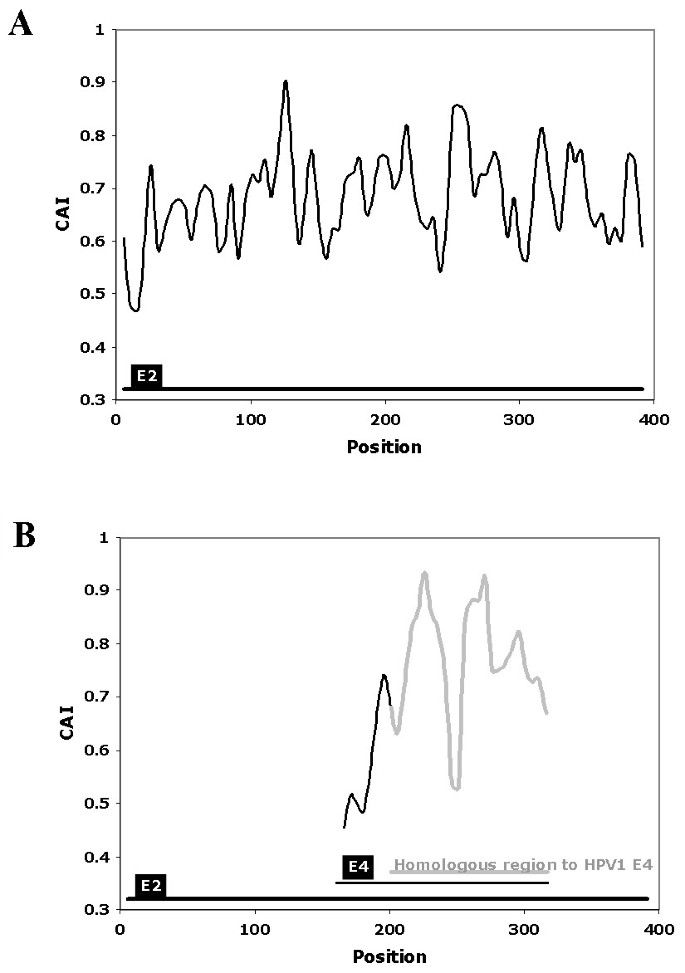
Codon adaptation index (CAI) (A), relative codon deoptimization index... | Download Scientific Diagram
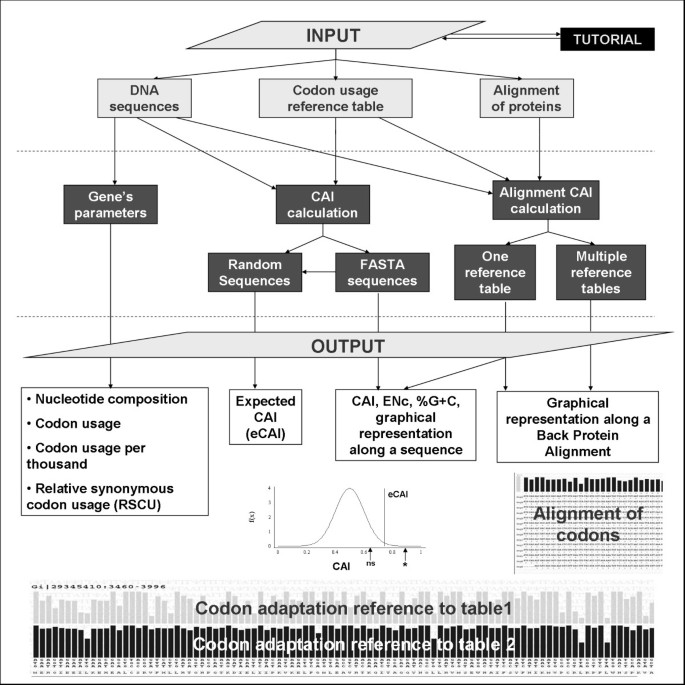
E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI) | BMC Bioinformatics | Full Text
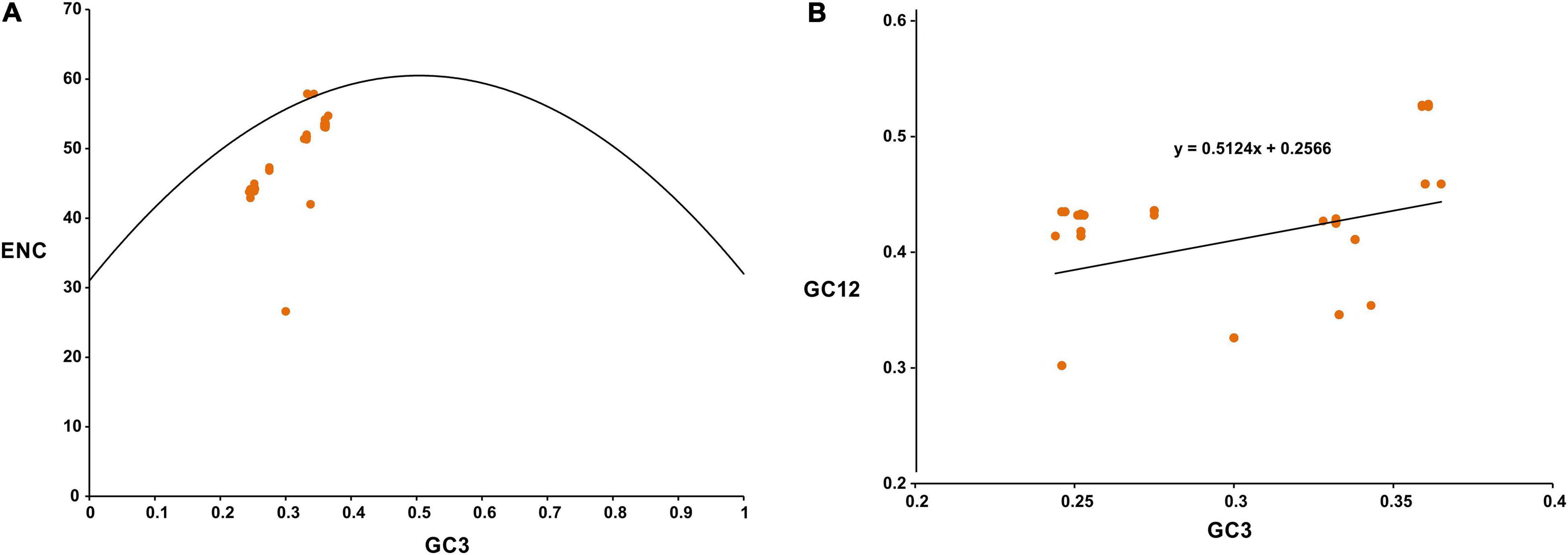
Frontiers | Base Composition and Host Adaptation of the SARS-CoV-2: Insight From the Codon Usage Perspective

Relative Codon Adaptation Index, a Sensitive Measure of Codon Usage Bias - Soohyun Lee, Seyeon Weon, Sooncheol Lee, Changwon Kang, 2010

A large-scale analysis of codon usage bias in 4868 bacterial genomes shows association of codon adaptation index with GC content, protein functional domains and bacterial phenotypes - ScienceDirect
COMPARATIVE ANALYSIS OF CODON USAGE PATTERNS AND IDENTIFICATION OF PREDICTED HIGHLY EXPRESSED GENES IN FIVE SALMONELLA GENOMES

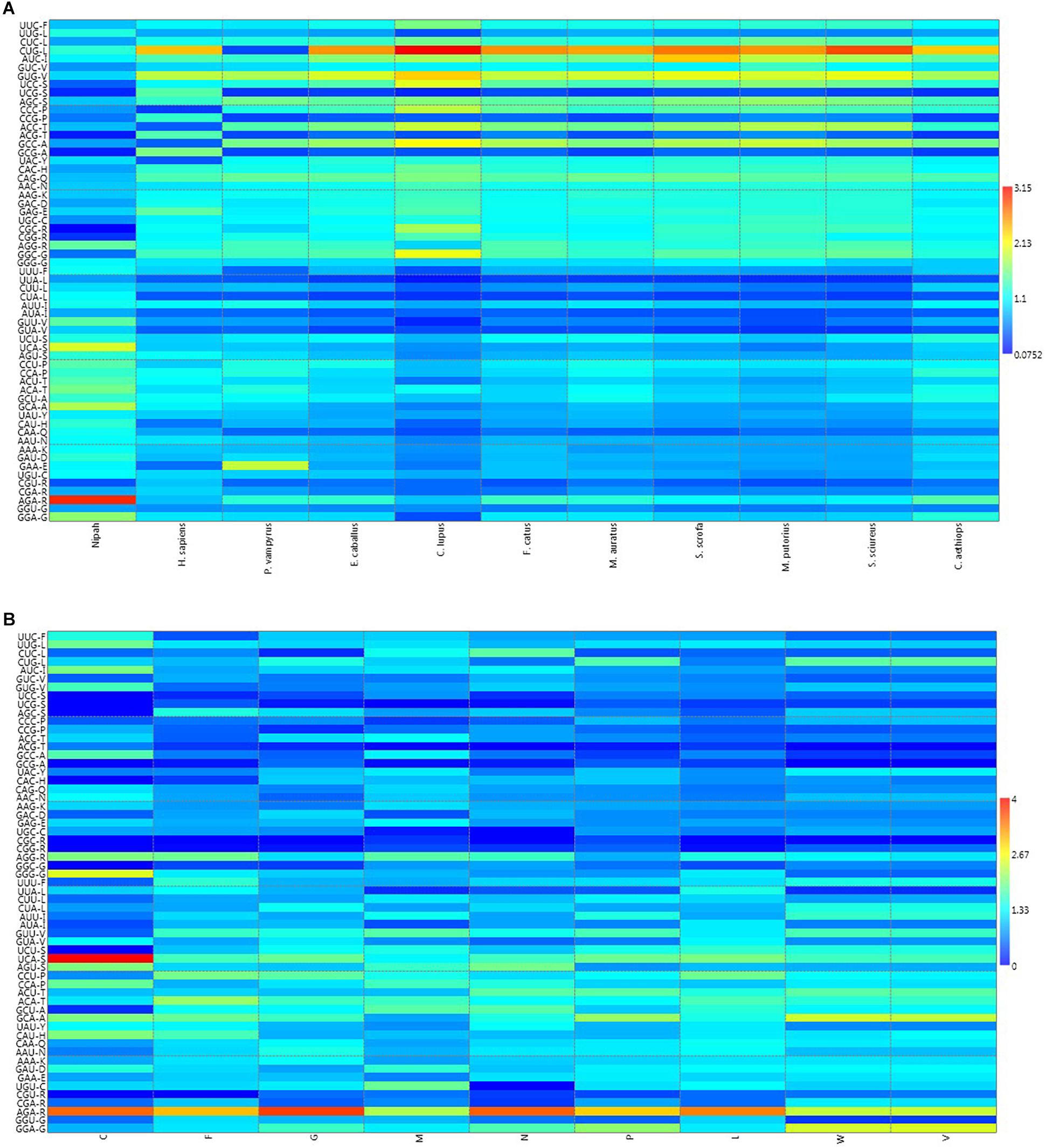



![Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ] Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2015/1520/1/fig-3-full.png)